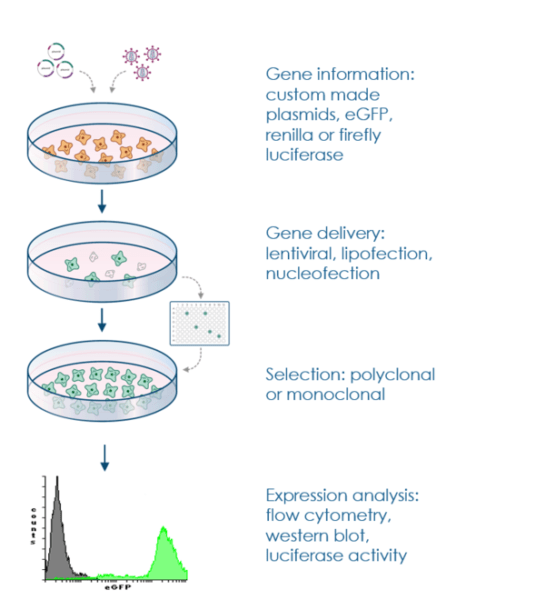
Stable cell lines play a crucial role in advancing scientific research, enabling prolonged and consistent gene expression for various applications. This article provides a comprehensive guide for researchers seeking to establish stable cell lines that constitutively express GFP-tagged proteins. The protocol outlined below ensures successful integration of the gene of interest into the cell’s genome and robust expression, enabling a wide range of experimental possibilities.
Stable cell lines serve as robust platforms facilitating the exploration of protein expression, localization, and functional dynamics within the cellular context. These specialized cell lines are crafted by introducing a gene of interest into host cells, selectively nurturing those that have seamlessly integrated the gene into their genetic blueprint. The objective is to cultivate a cellular population where a substantial proportion consistently showcases the fusion protein’s expression. This concerted effort enables researchers to delve deep into the intricate behaviors and interactions exhibited by the fusion protein.
Stable cell lines can be generated through two main approaches: episomal maintenance and direct integration into the genome. Episomal maintenance involves introducing a gene of interest into cells using vectors designed to be retained as episomes within the nucleus. Alternatively, direct integration integrates the gene of interest into the host cell’s genome. The latter method offers greater stability and is commonly preferred for long-term studies.
Depending on the scope of the experiment, several options are used for the generation of a stable cell line. A mixed population of drug resistant cells can be used directly for experimental analysis with the advantage of generating fast results, but also the disadvantage of dealing with an undefined and genetically mixed cell population. Another option is to generate a monoclonal cell line. In this method, it is necessary to dilute the resistant cells by plating in 96-well plates in such a way that culture as single and isolated cells. Subsequently, the cloning of single cell may be repeated several times to obtain 100% clonal purity. This culture method can be used for screening experiments or conduction studies by using a homogenous and defined cell system.
The successful creation of stable cell lines hinges on the use of a selection marker, which ensures that only cells containing the gene of interest are propagated. Selection markers are often resistance genes to antibiotics like puromycin, neomycin, DHFR, or glutamine synthetase. The presence of these markers allows for the survival and expansion of cells that have integrated the transgene.
In order to select stably-transfected cells, a selection marker must be co-expressed with the target protein. The marker gene could be on either the same plasmid vector or a second, co-transfected vector. There are a variety of systems for selecting transfected cells, including resistance to antibiotics puromycin, neomycin, DHFR, and glutamine synthetase. After gene transfer, cells are developed in medium containing the selective agent. Only those cells which have contained the drug resistant gene survive.
The culture conditions of the selected cell type are pivotal for generating stable cell lines. Following the supplier’s cell culture recommendations, such as those provided by ATCC, is crucial for optimal results. Passaging cells two days before the experiment and maintaining passage numbers below 30 are generally recommended.
Choose a cell line that is relevant to your research and has the appropriate characteristics for your study. Commonly used cell lines include HEK293, CHO, HeLa, and others. The choice of cell line depends on factors such as growth characteristics, transfection efficiency, and compatibility with your study.
Create a plasmid vector that contains the gene or protein of interest along with a selectable marker gene (e.g., neomycin, puromycin resistance). The selectable marker gene will allow you to identify and select cells that have successfully integrated the plasmid into their genome.
Transfect the cells with the plasmid using a suitable transfection method, such as calcium phosphate transfection, lipofection, electroporation, or viral transduction. This step introduces the plasmid DNA into the cells.
Add a selection agent (usually an antibiotic like neomycin or puromycin) to the culture media to kill off cells that have not incorporated the plasmid. This step selects for cells that have successfully integrated the plasmid and the selectable marker gene.
Once the transfected cells have survived the selection process and formed colonies, perform single-cell cloning. Dilute the cells to a low density so that each well or dish contains only a single cell. This will ensure that each resulting colony is derived from a single cell.
Expand the individual colonies into larger cultures. Test the expression of your gene or protein of interest using methods like Western blotting, immunofluorescence, or RT-qPCR. Select colonies that exhibit the desired expression levels for further characterization.
Perform functional assays or experiments to confirm that the stable cell line expresses the gene or protein of interest and behaves as expected. This may involve assessing biological activity, protein-protein interactions, or cellular responses.
Once you’ve confirmed the stability and functionality of your stable cell line, cryopreserve multiple vials of cells at early passages. This will ensure a backup in case of contamination or experimental mishaps.
Maintain detailed records of the entire process, including plasmid sequences, transfection protocols, selection conditions, single cell cloning procedures, and any assay results. This documentation is crucial for reproducibility and troubleshooting.
Regularly passage and maintain your stable cell line under appropriate culture conditions. This involves monitoring growth rates, expression levels, and phenotypic characteristics to ensure the stability and consistency of the cell line over time.

Stable cell lines have a wide range of applications in various fields of research and biotechnology due to their ability to express specific genes or proteins consistently. Here are some key applications of stable cell lines:
Establishing stable cell lines offers researchers a valuable tool for prolonged gene expression studies. By following this step-by-step protocol, researchers can confidently generate stable cell lines expressing GFP-tagged proteins. These cell lines provide a foundation for investigating protein behavior, interactions, and localization, contributing to advancements in cell biology and biomedical research. As technology evolves, stable cell lines continue to be instrumental in uncovering the intricacies of cellular processes.
Generating stable cell lines allows you to express specific genes or proteins of interest in a controlled and consistent manner. This enables you to study their functions, interactions, and regulation over time, providing valuable insights into cellular processes.
A selectable marker is a gene introduced into cells along with the gene of interest. It confers resistance to an antibiotic or other selective agent. Cells that successfully incorporate the selectable marker are able to survive in the presence of the selective agent, making it easier to isolate and maintain cells expressing your gene of interest.
Commonly used selection agents include antibiotics like neomycin, puromycin, and hygromycin. These agents inhibit the growth of cells that have not integrated the plasmid containing the selectable marker gene.
Choose a cell line that is relevant to your research and has characteristics suitable for your study (growth rate, transfection efficiency, origin, etc.). Ensure the chosen cell line is compatible with the experimental systems you plan to use.
Transfection efficiency can be optimized by testing different transfection methods, using appropriate ratios of plasmid DNA to transfection reagents, and considering factors like cell density and serum concentration in the culture medium.
Diluting the cells to a low density in the culture dish or well increases the likelihood that each well contains only one cell. Microscopy can help verify this. It’s important to use sterile techniques to prevent contamination during single-cell cloning.
Confirm plasmid integration through methods like PCR, Southern blotting, or fluorescence in situ hybridization (FISH) targeting the integrated gene. These methods help verify that the gene of interest has been successfully integrated into the genomic DNA.
Regularly monitor the expression of your gene or protein of interest through assays like Western blotting, immunofluorescence, or flow cytometry. Perform consistent passaging and maintain uniform culture conditions to prevent genetic and phenotypic changes.
Maintain cells in sterile conditions, monitor for contamination, and store multiple vials of cells at different passages in liquid nitrogen to ensure backup stocks. Maintain detailed records of passages and characteristics of the cells.
Troubleshoot by optimizing transfection conditions, selecting appropriate antibiotics, and verifying plasmid integration. If expression is low or unstable, check for genomic integration and consider evaluating different promoters or enhancers.
I'm Binod G C (MSc), a PhD candidate in cell and molecular biology who works as a biology educator and enjoys scientific blogging. My proclivity for blogging is intended to make notes and study materials more accessible to students. View all posts by Binod G C